AI-Powered Chest CT Reporting: Transforming Radiologist Observations into Standardized Data
How do we automatically transform the plain text of imaging reports into rich, actionable data? The process is far from simple. In this post, I’ll walk you through how we’re using artificial intelligence to make this happen—and how you can get involved if you’re interested!
The Core Problem: Standardizing the Observations within Radiology Reports
Radiology reports contain crucial information that guide patient management, but the language used can vary widely from one radiologist to another. This variability can make it challenging to share, understand, and use the data consistently. What we need is a universal language that standardizes the way these findings are described. Enter Common Data Elements.
What Are Common Data Elements (CDEs)?
CDEs provide a standardized framework to encode the detailed information found in radiology reports, linked with other standard ontologies (Anatomic Locations, RadLex, SNOMED CT). Think of them as a universal language for describing imaging findings. This consistency is key to developing and integrating clinical tools that can enhance patient care.
Imagine a patient, John, who arrives at the hospital with chest pain. A chest CT scan is performed, and the radiologist incidentally notes calcifications in John’s coronary arteries. These calcifications are a significant finding that can lead to heart disease. What should we expect to happen next? Ideally, this finding would encoded into a standardized message and sent to his electronic medical record (EMR). The EMR would then trigger an alert for John’s primary care physician, recommending further evaluation and follow-up actions to address potential heart disease.
Our Brand New AI-Powered Approach to Creating CDEs
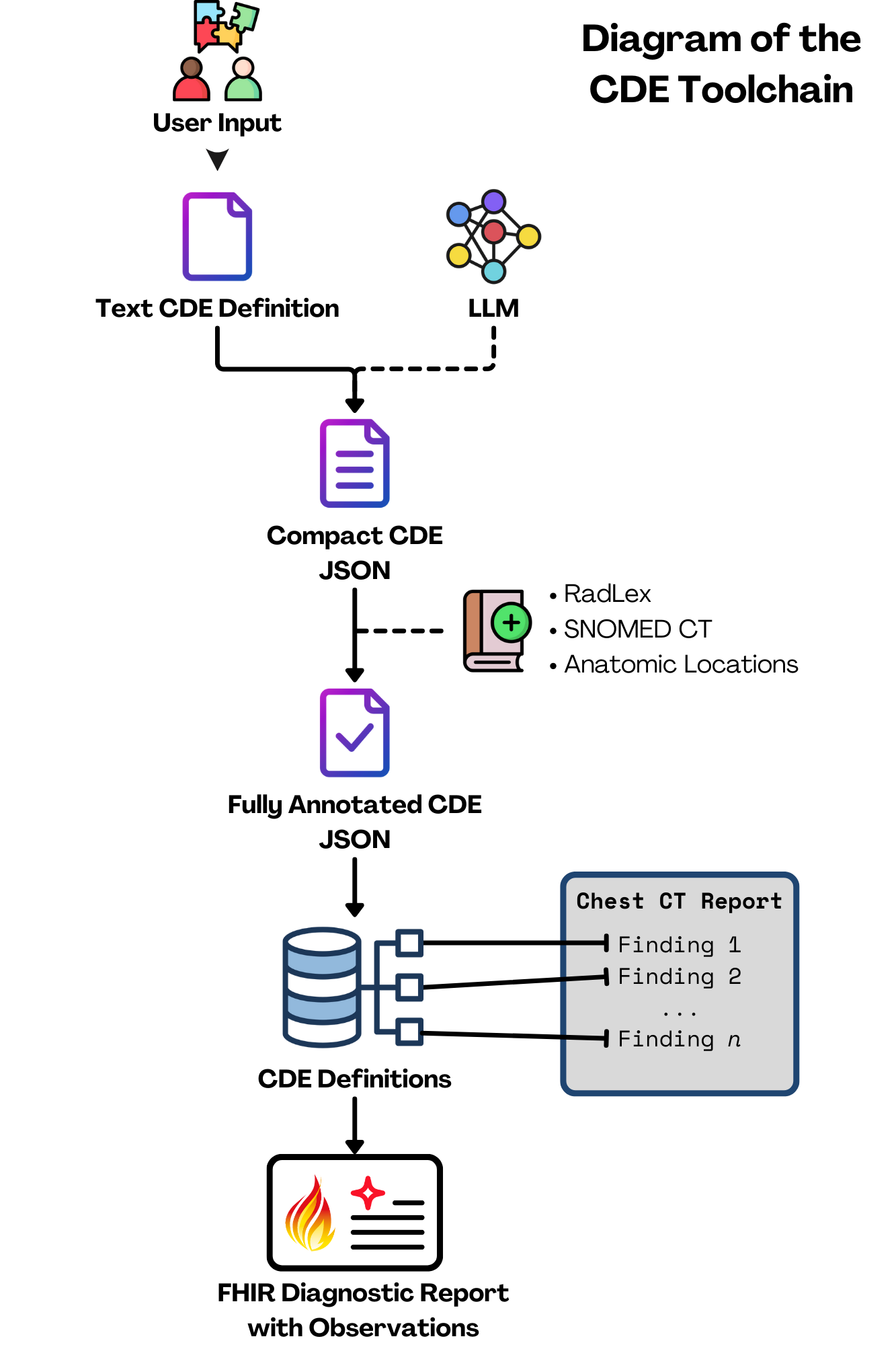
Now, you might be thinking, “Radiologists make a ton of different observations. Just how many CDEs are we talking about here?” It's as many as you'd imagine. Thousands of CDEs would be required to describe every finding, for each imaging modality. Creating these manually for every possible finding is an absolutely enormous task. To tackle this challenge, we chose an initial scope of chest CTs and developed a novel approach that uses a large language model (LLM) to assist in generating these CDEs. Here’s a simplified look at how it works:
- Gathering the Data:
- We started by collecting a large set of anonymized chest CT reports.
- Each report was broken down into smaller, overlapping sections, making it easier to analyze.
- Understanding the Data:
- These sections were converted into semantic vectors using an embedding model. Think of these vectors as unique fingerprints that capture the essence of each text chunk.
- These vectors were stored in a database that allows for quick and efficient searching.
- Finding Relevant Information:
- To create a CDE for a specific finding (like a pulmonary nodule), the tool searches the database of CT reports for text chunks similar to the finding name.
- It uses a cross-encoder to rank these chunks by relevance, ensuring the most pertinent information is prioritized.
- Generating the CDEs:
- The top-ranked chunks are fed into a GPT model, which generates a preliminary CDE.
- This model describes the finding in detail, including its presence, characteristics (like size and location), and in our first iteration of this concept, any associated observations that describe adjacent or relevant anatomy that isn't the actual finding itself.
- Review and Polish:
- After the AI does its job, a radiologist and/or team of content experts reviews each CDE to ensure it's clinically accurate and useful.
- Further annotations are applied to the CDE (including the relevant ontological codes from Anatomic Locations, RadLex, and SNOMED) to create a CDE set definition.
Pilot Testing and Real-World Application
We put our method to the test with an initial pilot of the most common findings in chest CT reports. This iterative process helped us refine and define CDEs for 200+ different findings, which are now available on GitHub for further review.
The Big Picture: Why This Matters
By using AI to standardize radiology reporting, we both make the job easier for radiologists and also improve the accuracy and consistency of patient data. This leads to better diagnostics, more efficient care, and ultimately, improved patient outcomes.
In our example of John's coronary arteries, this might look like the following:
- Initial observation:
- The radiologist identifies coronary artery calcifications on John’s chest CT scan.
- This finding is recorded in the radiology report, noting the presence, severity, and potentially other features such as the specific location and/or density of the calcifications.
- Automated message to the EMR:
- The observation is encoded into a CDE.
- This CDE is formatted into a Fast Healthcare Interoperability Resources (FHIR) message, an interoperability standard.
- The FHIR message is sent to John’s EMR and to downstream clinical tools, such as an automated tool that might calculate John's Agatston score (which can help stratify risk for a major adverse cardiac event).
- Triggering clinical decision support (CDS):
- Upon receiving the FHIR message, the EMR automatically triggers a CDS tool.
- The CDS tool alerts John’s primary care physician about the significant finding.
- The alert includes recommendations for risk stratification and suggesting further evaluation by a cardiologist.
- The CDS tool may also prompt specific follow-up interventions, like an order for a cardiology consultation and or additional diagnostic tests if necessary.
Join Us in Revolutionizing Radiology
Are you interested in imaging informatics or AI in healthcare? We invite you to join us on this journey! Whether you’re a radiologist, a resident/fellow, a student, a data scientist, or just someone passionate about imaging informatics, there’s a place for you to contribute. Check out our upcoming SIIM 2024 Hackathon Project, GitHub repository, provide feedback, and collaborate with us to help expand and refine this essential work.
Stay tuned as we continue to refine our approach and expand our scope to other anatomic locations. We're excited to be at the forefront of this revolution, and we have many more exciting developments ahead.
Don't forget to subscribe to OIDM for the next update.

