Attaching Ontology Links to Common Data Elements
The Problem
One of the biggest challenges in the creation of Common Data Elements (CDEs) is the integration with different ontologies. Each system—whether it’s SNOMED, RadLex, or Anatomic Locations—organizes medical concepts differently, often with overlapping codes and varying terminologies. As discussed in the last post, thousands of CDEs are needed for our goal of transforming the free text radiology reports into structured, actionable data. However, the manual selection of these codes is time-consuming and prone to errors. So, how do we solve this?
The Goal: Automated CDE → Ontology Mapping
Ontologies in the medical field are like specialized dictionaries that organize and define medical terms and concepts. Imagine you have different dictionaries for the same language, but each one organizes words differently and uses slightly different definitions. In medicine, these dictionaries are called ontologies, and they include systems like SNOMED, RadLex, and Anatomic Locations.
Each ontology categorizes medical information—like symptoms, diagnoses, and procedures—so doctors and computers can understand and use the data effectively. However, because each ontology has its own way of organizing and defining terms, it can be tricky to ensure that everyone is on the same page. That’s why we need to match these ontologies to CDEs. This matching helps ensure that no matter which ontology is used, the medical information is consistent and can be accurately shared and understood across different healthcare applications.
Currently, the process of matching the right codes to CDEs representing clinical findings is done manually. It’s time-consuming, prone to errors, and can be inconsistent. We manually select the best-matched codes for each CDE from the available terminologies, using code lookups to find the most appropriate codes from each ontology that represent specific observations. By meticulously curating these examples, we aim to teach our automated CDE tool to replicate this process autonomously, selecting the best codes for all other CDEs.
Example/Workflow
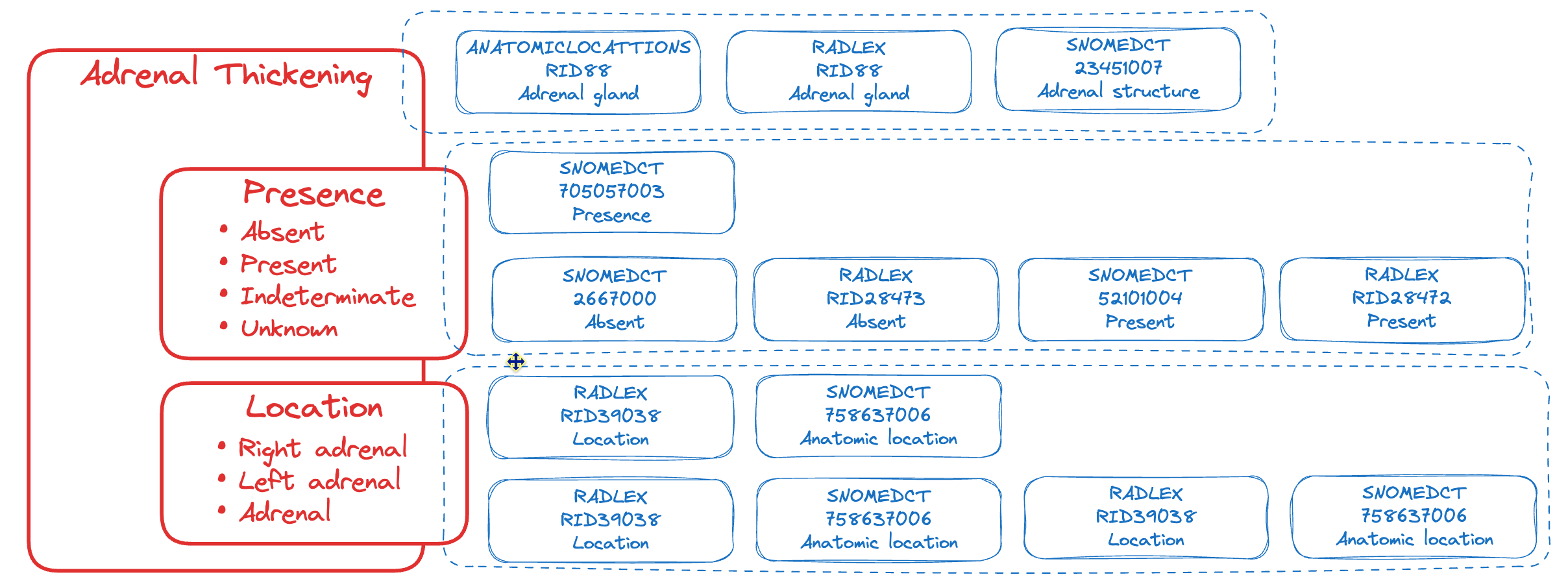
Let’s break it down and consider an example. Using our model, we search for matching SNOMED, RadLex, and Anatomical Locations codes. These codes are then linked to the CDE, providing a standardized and interoperable representation of the finding. The figure above demonstrates this mapping process, showcasing how a sample of codes corresponds to a specific aspect of the CDE.
Adrenal gland thickening is evidenced by increased uniform or focal gland thickness on CT or MRI, with the normal limb width typically less than 10 mm; imaging features such as attenuation values, enhancement patterns, and presence of mass effect may assist in differentiating between benign and malignant etiologies.
Obviously, we'd carry forward and incorporate many more codes from all of the ontologies at all the levels: the finding itself (CDE Set), each attribute (CDE Element), and attribute values. These values can then be included with a radiologist's description of a finding when it's encoded using CDEs.
Come to the SIIM Hackathon
We'll be in National Harbor at SIIM next week, and we're working on making at least a demonstration of how this might work in practice. Come see and offer your thoughts/feedback!
